-Search query
-Search result
Showing 1 - 50 of 518 items for (author: singh & v)

EMDB-44254: 
Consensus map: CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-42628: 
Structure of the insect gustatory receptor Gr9 from Bombyx mori
Method: single particle / : Gomes JV, Butterwick JA

EMDB-42629: 
Structure of the insect gustatory receptor Gr9 from Bombyx mori in complex with D-fructose
Method: single particle / : Gomes JV, Butterwick JA

EMDB-43548: 
Structure of the insect gustatory receptor Gr9 from Bombyx mori in complex with L-sorbose
Method: single particle / : Gomes JV, Butterwick JA

PDB-8uvt: 
Structure of the insect gustatory receptor Gr9 from Bombyx mori
Method: single particle / : Gomes JV, Butterwick JA

PDB-8uvu: 
Structure of the insect gustatory receptor Gr9 from Bombyx mori in complex with D-fructose
Method: single particle / : Gomes JV, Butterwick JA

PDB-8vv3: 
Structure of the insect gustatory receptor Gr9 from Bombyx mori in complex with L-sorbose
Method: single particle / : Gomes JV, Butterwick JA

EMDB-35817: 
Structure of Niacin-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-35822: 
Structure of MK6892-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-35831: 
Structure of GSK256073-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-36193: 
Structure of Acipimox-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-36280: 
Structure of MMF-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8iy9: 
Structure of Niacin-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8iyh: 
Structure of MK6892-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8iyw: 
Structure of GSK256073-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8jer: 
Structure of Acipimox-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8jhn: 
Structure of MMF-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-43256: 
CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-43258: 
CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-43327: 
CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-43328: 
CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

PDB-8vib: 
CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

PDB-8vid: 
CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

PDB-8vkq: 
CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

PDB-8vkr: 
CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-41100: 
CCW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming 34-mer C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-41101: 
33-mer FliF MS-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-41102: 
33-mer Flagellar Switch Complex - FliF, FliG, FliM and FliN forming C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-41103: 
35-mer Flagellar Switch Complex - FliF, FliG, FliM and FliN forming C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

EMDB-41104: 
36-mer Flagellar Switch Complex - FliF, FliG, FliM and FliN forming C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

PDB-8t8o: 
CCW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming 34-mer C-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM

PDB-8t8p: 
33-mer FliF MS-ring from Salmonella
Method: single particle / : Singh PK, Iverson TM
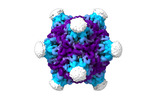
EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
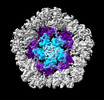
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
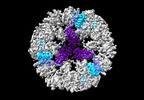
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
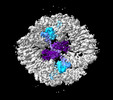
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb3: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb4: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb5: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb7: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-16407: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (focussed on SSU head)
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16408: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (focussed on SSU body)
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16409: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (masked refined on SSU tail)
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16410: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (focussed on LSU body)
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16411: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (focussed on L10/L12 stalk)
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16412: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (focussed on mS39-LRPPRC-SLIRP)
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16413: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (focussed on central protuberance)
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16414: 
Human mitochondrial ribosome in complex with LRPPRC-SLIRP, A-site, P-site, E-site tRNAs and mRNA (consensus)
Method: single particle / : Singh V, Itoh Y, Amunts A
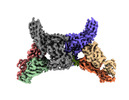
EMDB-34174: 
Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Atypical chemokine receptor 2, ACKR2 (D6R)
Method: single particle / : Maharana J, Sarma P, Yadav MK, Banerjee R, Shukla AK
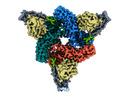
EMDB-36078: 
Structure of beta-arrestin2 in complex with M2Rpp
Method: single particle / : Maharana J, Sano FK, Shihoya W, Banerjee R, Nureki O, Shukla AK
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
